Use colony PCR to identify the correct clones
(40pts) 1. As a Bio195/196 student your project is to clone the promoter region of mRMRP into a plasmid vector. You plan to use colony PCR to identify the correct clones but you need to design primers. You put the sequence into the primer design software and the following primer sequences were identified.
Parameters of primer selection: Sequence Scanned: 825 to 980
Found 24 primer pairs
Analysis settings:
primer size: 18 - 25 3' dinucleotide: NS
Tm (¡C): 45 - 80 percent G+C content: 40 - 60
product size: 100 - 156
Maximum consecutive bonds allowed:
primer vs. primer (any): 5 3'-end vs. 3'-end: 4
primer vs. primer (G-C only): 5 3'-end vs. product: 4
[Forward F1] 825-842 5'- TCGCGCCTAGAAGTTAAG -3'
18 nt forward primer
pct G+C: 50.0 Tm: 45.8
[Backward B1] 980-956 5'- AGTGACTTGCGGGGGAAGTCTACGC -3'
25 nt backward primer
pct G+C: 60.0 Tm: 64.3
[F2] 829-847 5'- GCCTAGAAGTTAAGGCAGC -3'
19 nt forward primer
pct G+C: 52.6 Tm: 46.2
[B1] 980-956 5'- AGTGACTTGCGGGGGAAGTCTACGC -3'
25 nt backward primer
pct G+C: 60.0 Tm: 64.3
[F3] 834-851 5'- GAAGTTAAGGCAGCTCGC -3'
18 nt forward primer
pct G+C: 55.6 Tm: 47.5
[B3] 976-958 5'- ACTTGCGGGGGAAGTCTAC -3'
19 nt backward primer
pct G+C: 57.9 Tm: 50.6
[F6] 836-853 5'- AGTTAAGGCAGCTCGCTC -3'
18 nt forward primer
pct G+C: 55.6 Tm: 47.0
[B3] 976-958 5'- ACTTGCGGGGGAAGTCTAC -3'
19 nt backward primer
pct G+C: 57.9 Tm: 50.6
- Choose a primer pair and justify your choice.
- Determine the size of the PCR amplicon produced.
- Specify what annealing temperature you would use.
The first time you run a reaction a non-specific PCR product is amplified in addition to the correct amplicon. What would you do to troubleshoot the assay?
(30pts) 2. You have just joined a lab and are given the following project. The lab has been growing cancer cell lines in the presence of gamma tocopherol and has shown that it causes the cancer cells to undergo apoptosis (programmed cell death). The lab would like to identify which genes are involved. Design a project to identify which genes might be involved so that you can impress the PI and get a good recommendation for med school. How will you confirm your initial results? Be sure to detail your experimental design.
(30pts) 3. Given the following quantitative reverse transcriptase-real time PCR experimental data draw a standard curve and determine the copy number of the unknowns. If sample 231 is the initial sample and 278 is the sample after treatment what is the log fold difference between the samples? Is this experiment well designed? Why or why not?
|
Well |
Sample Name |
Detector Name |
Reporter |
Task |
Ct |
Quantity |
|
3 |
278 |
PML-RARA |
FAM |
Unknown |
28.67186 |
? |
|
15 |
278 |
PML-RARA |
FAM |
Unknown |
28.90929 |
? |
|
27 |
231 |
PML-RARA |
FAM |
Unknown |
25.87421 |
? |
|
39 |
231 |
PML-RARA |
FAM |
Unknown |
25.76226 |
? |
|
74 |
NTC |
PML-RARA |
FAM |
Unknown |
Undetermined |
0 |
|
86 |
NTC |
PML-RARA |
FAM |
Unknown |
Undetermined |
0 |
|
1 |
SP1 |
PML-RARA |
FAM |
Standard |
38.52142 |
10 |
|
13 |
SP1 |
PML-RARA |
FAM |
Standard |
39.71262 |
10 |
|
25 |
SP2 |
PML-RARA |
FAM |
Standard |
34.62336 |
100 |
|
49 |
SP3 |
PML-RARA |
FAM |
Standard |
30.99905 |
1000 |
|
61 |
SP3 |
PML-RARA |
FAM |
Standard |
31.3191 |
1000 |
|
73 |
SP5 |
PML-RARA |
FAM |
Standard |
24.01704 |
100000 |
|
85 |
SP5 |
PML-RARA |
FAM |
Standard |
24.23932 |
100000 |
|
2 |
SP6 |
PML-RARA |
FAM |
Standard |
20.61799 |
1000000 |
|
14 |
SP6 |
PML-RARA |
FAM |
Standard |
20.4106 |
1000000 |
(30pts) 4. Briefly describe the 2 main high throughput NGS sequencing systems (Illumina HiSEQ/MiSEQ and Life Technologies Ion Torrent/PROTON) and identify the advantages and disadvantages of each. What general category of applications would be most appropriate for the MiSEQ/Ion Torrent vs the HiSEQ/Proton instruments?
(30 pts) 5. A-What are the three types of components combined in a flow cytometer?
B-What type of light source is used in most flow cytometers?
C- What types of information can be obtained about a cell, using flow cytometry?
D- Delineate how a single laser line (eg, 488 nm) can be used in conjunction with multiple fluorescent dyes of varying emission wavelengths. Define the term “Stokes Shift”
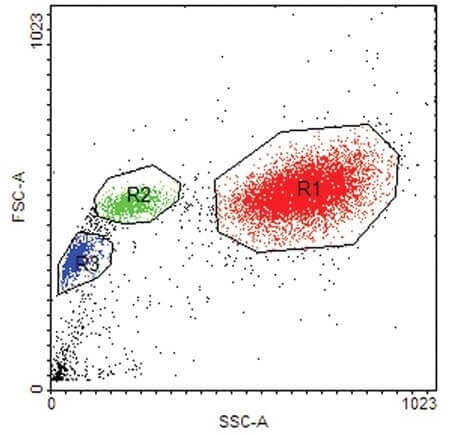
(20 pts) 6. What can be derived from the scatter plot below with respect to the circled groups of cells- R1, R2 and R3?
(20pts) Q.7A Why can’t all samples be analyzed by GC/MS?
B What is the basis for separation in MS?
C What is accurate mass?
D What is the purpose of quadrupole rods?
E For what type of samples would MALDI_TOF be the method of choice?
(50pts) Q.8.We have five patients with different tumor types and they were sequenced on MiSeq. Please see the list of patients and variants found below:
Patient 1: Colorectal cancer
AKT1 p. E17K
EGFR p.T259T
Patient 2: Non-Small Cell Lung Cancer
EGFR p.G719A
PDGFRA p.P567P
Patient 3: Glioma
EGFR p.V292M
IDH1 p.R132H
Patient 4: Breast Cancer
PTEN p.R233*
NRAS p.A59A
Patient 5: Acute Lymphoblastic Leukemia
KRAS p. A59G
JAK2 p.R683G
- Prepare a table with the information below for each variant:
Coordinates in hg19
Nucleotide change
Exon number where the variant is
SNP rs number
COSMIC ID
Frequency of the mutation in the cancer type
Clinical significance
Response to therapies
- Based on the information you found on these variants, which ones are significant, which ones are artifacts? Why?
- List all databases you used.
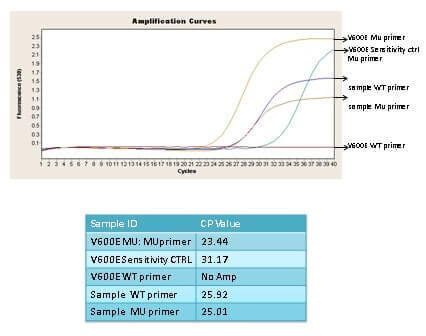
(20 pts) Q 9. Given the real time pcr curve below from a MASA experiment does the sample have a BRAF V600E mutation? Explain your reasoning. What is the 3” nucleotide of the WT and Mutant primer? If the sample had a BRAF V600M mutation would this MASA assay detect it?
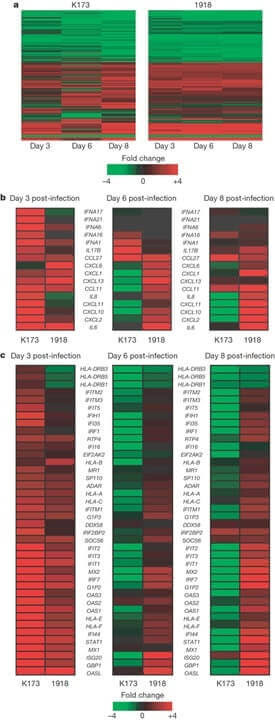
(30 pts) Q.10 Shown below is a microarray experiment comparing 2 cell lines undergoing treatment, where the fold change in expression is shown by the key. What is this type of data representation called? In panel B which cytokine or chemokine genes are changing in expression between the 3 time points? What about between the 2 cell lines at each time point?
Panel C compares the expression of type I interferon (IFN)-stimulated genes, which time point shows the greatest difference between K173 and 1918? You join the lab at this point to continue the project what would be your next step?